Dipartimento di Matematica Guido Castelnuovo, Università Sapienza Roma
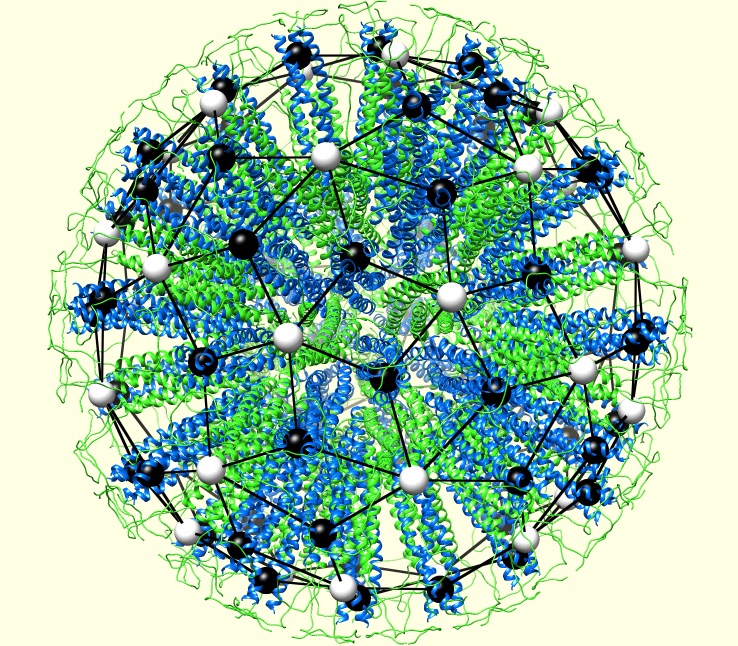
Abstract: Several nanoparticles self-assemble from multiple copies of a limited number of building blocks, viruses being the archetypal example in biology. A majority of viruses have a highly ordered protein container (the capsid) that encloses and protects the viral genomic material from the environment. Understanding the structure of these capsids is fundamental to clarify how viruses work and possibly how they can be defeated and, to this purpose, a general classification scheme is needed. For viruses, the theory that identifies the relative position of the proteins was proposed by Caspar and Klug in the sixties and is still the current paradigm in the description of capsids. However, in the last decade scientists have started engineering many different self-assembling protein nanoparticles. The possible applications are manifold and innovative: from vectors for the delivery of genes or drugs to synthetic vaccines. The fast development of this research field poses new challenges for the structural analysis as, in general, synthetic nanoparticles do not fit into the Caspar and Klug’s framework: an original approach is needed to provide blueprints for the experimental determination of their structure. We focus on a de novo class of proteins, referred to as self-assembling protein nanoparticles (SAPNs). This is a family of nanoparticles that provide a versatile platform for synthetic vaccines against a broad range of diseases, including malaria, SARS, influenza and HIV. These nanoparticles self assemble from multiple copies of a polypeptide synthesized to have specific connectivity properties. The polypeptides bind to each other following precise local assembly rules forming groups of five or three polypeptides. To address the problem of elucidating the resulting assembled global structure, we use tools from graph theory. This approach unveils a hidden relation with fullerene geometries and enables a full classification of the high and low symmetry particles seen in experiments. First and foremost, this allows to determine the relative distance of the epitopes on the particle surface and tune the immunogenic effect of the vaccine.